Tutorial 5: 3D spatial domain identification
We extended STAGATE for 3D spatial domain identification by simultaneously considering the 2D SNN within each section and neighboring spots between adjacent section. The key idea of the usage of 3D SNN is that the biological differences between consecutive sections should be continuous, so we can enhance the similarity between adjacent sections to eliminate the discontinuous independent technical noises.
In this tutorial, we applied STAGATE onto a pseudo 3D ST data constructed by aligning the spots of the “cord-like” structure in seven hippocampus sections profiled by Slide-seq.
The original Slide-seq can be downloaded from https://portals.broadinstitute.org/single_cell/study/slide-seq-study. We provided the processed data and aligned coordinates at https://drive.google.com/drive/folders/10lhz5VY7YfvHrtV40MwaqLmWz56U9eBP?usp=sharing.
Preparation
[1]:
import warnings
warnings.filterwarnings("ignore")
[2]:
import pandas as pd
import numpy as np
import scanpy as sc
import matplotlib.pyplot as plt
import os
import sys
[3]:
import STAGATE
[4]:
data = pd.read_csv('Data/3D_Hippo_expression.txt', sep='\t', index_col=0)
Aligned_coor = pd.read_csv('Data/ICP_Align_Coor.txt', sep='\t', index_col=0)
[5]:
Aligned_coor.head()
[5]:
| X | Y | Z | Section | |
|---|---|---|---|---|
| AAAAACAACCAAT-13 | 3946.072727 | 3599.436364 | 6000 | Puck_180531_13 |
| AAAAGTCCATTAT-13 | 2401.628788 | 3550.333333 | 6000 | Puck_180531_13 |
| AAACAACGCGCGA-13 | 2921.968153 | 3353.687898 | 6000 | Puck_180531_13 |
| AAACAATTCAATA-13 | 2711.556338 | 3365.542254 | 6000 | Puck_180531_13 |
| AAACACGCTGCCC-13 | 2351.863354 | 4265.447205 | 6000 | Puck_180531_13 |
[6]:
adata = sc.AnnData(data)
adata
[6]:
AnnData object with n_obs × n_vars = 10908 × 9420
[7]:
#Normalization
sc.pp.highly_variable_genes(adata, flavor="seurat_v3", n_top_genes=3000)
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
[8]:
# loading metadata and aligned coordinates
adata.obs['X'] = Aligned_coor.loc[adata.obs_names, 'X']
adata.obs['Y'] = Aligned_coor.loc[adata.obs_names, 'Y']
adata.obs['Z'] = Aligned_coor.loc[adata.obs_names, 'Z']
adata.obs['Section_id'] = Aligned_coor.loc[adata.obs_names, 'Section']
[9]:
# loading the spatial locations
adata.obsm['spatial'] = adata.obs.loc[:, ['X', 'Y']].values
[10]:
section_colors = ['#02899A', '#0E994D', '#86C049', '#FBB21F', '#F48022', '#DA5326', '#BA3326']
[11]:
fig = plt.figure(figsize=(4, 4))
ax1 = plt.axes(projection='3d')
for it, label in enumerate(np.unique(adata.obs['Section_id'])):
temp_Coor = adata.obs.loc[adata.obs['Section_id']==label, :]
temp_xd = temp_Coor['X']
temp_yd = temp_Coor['Y']
temp_zd = temp_Coor['Z']
ax1.scatter3D(temp_xd, temp_yd, temp_zd, c=section_colors[it],s=0.2, marker="o", label=label)
ax1.set_xlabel('')
ax1.set_ylabel('')
ax1.set_zlabel('')
ax1.set_xticklabels([])
ax1.set_yticklabels([])
ax1.set_zticklabels([])
plt.legend(bbox_to_anchor=(1,0.8), markerscale=10, frameon=False)
ax1.elev = 45
ax1.azim = -20
plt.show()
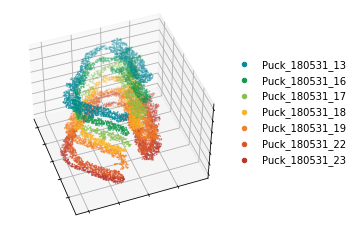
Constructing 3D spatial networks
[12]:
section_order = ['Puck_180531_13', 'Puck_180531_16', 'Puck_180531_17',
'Puck_180531_18', 'Puck_180531_19', 'Puck_180531_22',
'Puck_180531_23']
[13]:
STAGATE.Cal_Spatial_Net_3D(adata, rad_cutoff_2D=50, rad_cutoff_Zaxis=50,
key_section='Section_id', section_order = section_order, verbose=True)
Radius used for 2D SNN: 50
Radius used for SNN between sections: 50
------Calculating 2D SNN of section Puck_180531_13
Trying to set attribute `.uns` of view, copying.
This graph contains 22974 edges, 1692 cells.
13.5780 neighbors per cell on average.
------Calculating 2D SNN of section Puck_180531_16
Trying to set attribute `.uns` of view, copying.
This graph contains 20546 edges, 1476 cells.
13.9201 neighbors per cell on average.
------Calculating 2D SNN of section Puck_180531_17
Trying to set attribute `.uns` of view, copying.
This graph contains 11794 edges, 1138 cells.
10.3638 neighbors per cell on average.
------Calculating 2D SNN of section Puck_180531_18
Trying to set attribute `.uns` of view, copying.
This graph contains 18278 edges, 1363 cells.
13.4101 neighbors per cell on average.
------Calculating 2D SNN of section Puck_180531_19
Trying to set attribute `.uns` of view, copying.
This graph contains 22344 edges, 1788 cells.
12.4966 neighbors per cell on average.
------Calculating 2D SNN of section Puck_180531_22
Trying to set attribute `.uns` of view, copying.
This graph contains 24544 edges, 1835 cells.
13.3755 neighbors per cell on average.
------Calculating 2D SNN of section Puck_180531_23
Trying to set attribute `.uns` of view, copying.
This graph contains 18922 edges, 1616 cells.
11.7092 neighbors per cell on average.
------Calculating SNN between adjacent section Puck_180531_13 and Puck_180531_16.
Trying to set attribute `.uns` of view, copying.
This graph contains 34482 edges, 3168 cells.
10.8845 neighbors per cell on average.
------Calculating SNN between adjacent section Puck_180531_16 and Puck_180531_17.
Trying to set attribute `.uns` of view, copying.
This graph contains 23078 edges, 2614 cells.
8.8286 neighbors per cell on average.
------Calculating SNN between adjacent section Puck_180531_17 and Puck_180531_18.
Trying to set attribute `.uns` of view, copying.
This graph contains 25284 edges, 2501 cells.
10.1096 neighbors per cell on average.
------Calculating SNN between adjacent section Puck_180531_18 and Puck_180531_19.
Trying to set attribute `.uns` of view, copying.
This graph contains 24944 edges, 3151 cells.
7.9162 neighbors per cell on average.
------Calculating SNN between adjacent section Puck_180531_19 and Puck_180531_22.
Trying to set attribute `.uns` of view, copying.
This graph contains 26128 edges, 3623 cells.
7.2117 neighbors per cell on average.
------Calculating SNN between adjacent section Puck_180531_22 and Puck_180531_23.
Trying to set attribute `.uns` of view, copying.
This graph contains 21110 edges, 3451 cells.
6.1171 neighbors per cell on average.
3D SNN contains 294428 edges, 10908 cells.
26.9919 neighbors per cell on average.
Running STAGATE with 3D spatial networks
[14]:
adata = STAGATE.train_STAGATE(adata, alpha=0)
Size of Input: (10908, 3000)
WARNING:tensorflow:From /home/xzhou/anaconda3/envs/tensorflow/lib/python3.7/site-packages/tensorflow_core/python/ops/math_grad.py:1375: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.where in 2.0, which has the same broadcast rule as np.where
2022-04-29 21:26:31.725355: I tensorflow/core/platform/cpu_feature_guard.cc:142] Your CPU supports instructions that this TensorFlow binary was not compiled to use: AVX2 AVX512F FMA
2022-04-29 21:26:31.748905: I tensorflow/core/platform/profile_utils/cpu_utils.cc:94] CPU Frequency: 3499910000 Hz
2022-04-29 21:26:31.750571: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x55cb33581890 initialized for platform Host (this does not guarantee that XLA will be used). Devices:
2022-04-29 21:26:31.750614: I tensorflow/compiler/xla/service/service.cc:176] StreamExecutor device (0): Host, Default Version
2022-04-29 21:26:31.754849: I tensorflow/stream_executor/platform/default/dso_loader.cc:44] Successfully opened dynamic library libcuda.so.1
2022-04-29 21:26:32.055109: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x55cb35070bb0 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:
2022-04-29 21:26:32.055162: I tensorflow/compiler/xla/service/service.cc:176] StreamExecutor device (0): NVIDIA GeForce RTX 3090, Compute Capability 8.6
2022-04-29 21:26:32.055175: I tensorflow/compiler/xla/service/service.cc:176] StreamExecutor device (1): NVIDIA GeForce RTX 3090, Compute Capability 8.6
2022-04-29 21:26:32.057154: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1639] Found device 0 with properties:
name: NVIDIA GeForce RTX 3090 major: 8 minor: 6 memoryClockRate(GHz): 1.695
pciBusID: 0000:65:00.0
2022-04-29 21:26:32.057729: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1639] Found device 1 with properties:
name: NVIDIA GeForce RTX 3090 major: 8 minor: 6 memoryClockRate(GHz): 1.695
pciBusID: 0000:b3:00.0
2022-04-29 21:26:32.057850: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcudart.so.10.0'; dlerror: libcudart.so.10.0: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.057894: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcublas.so.10.0'; dlerror: libcublas.so.10.0: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.057927: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcufft.so.10.0'; dlerror: libcufft.so.10.0: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.057959: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcurand.so.10.0'; dlerror: libcurand.so.10.0: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.057992: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcusolver.so.10.0'; dlerror: libcusolver.so.10.0: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.058026: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcusparse.so.10.0'; dlerror: libcusparse.so.10.0: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.058057: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcudnn.so.7'; dlerror: libcudnn.so.7: cannot open shared object file: No such file or directory
2022-04-29 21:26:32.058061: W tensorflow/core/common_runtime/gpu/gpu_device.cc:1662] Cannot dlopen some GPU libraries. Please make sure the missing libraries mentioned above are installed properly if you would like to use GPU. Follow the guide at https://www.tensorflow.org/install/gpu for how to download and setup the required libraries for your platform.
Skipping registering GPU devices...
2022-04-29 21:26:32.058076: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1180] Device interconnect StreamExecutor with strength 1 edge matrix:
2022-04-29 21:26:32.058080: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1186] 0 1
2022-04-29 21:26:32.058083: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1199] 0: N N
2022-04-29 21:26:32.058086: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1199] 1: N N
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 500/500 [08:42<00:00, 1.05s/it]
[15]:
sc.pp.neighbors(adata, use_rep='STAGATE')
sc.tl.umap(adata)
[16]:
num_cluster = 4
adata = STAGATE.mclust_R(adata, num_cluster, used_obsm='STAGATE')
R[write to console]: Package 'mclust' version 5.4.7
Type 'citation("mclust")' for citing this R package in publications.
[17]:
adata.uns['Section_id_colors'] = ['#02899A', '#0E994D', '#86C049', '#FBB21F', '#F48022', '#DA5326', '#BA3326']
[18]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.umap(adata, color=['mclust', 'Section_id'])
... storing 'Section_id' as categorical
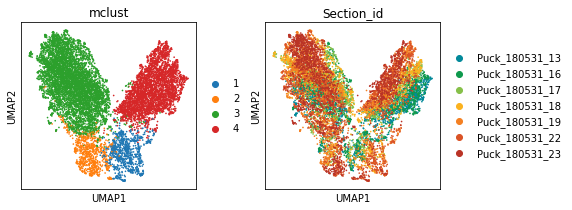
[19]:
fig = plt.figure(figsize=(4, 4))
ax1 = plt.axes(projection='3d')
for it, label in enumerate(np.unique(adata.obs['mclust'])):
temp_Coor = adata.obs.loc[adata.obs['mclust']==label, :]
temp_xd = temp_Coor['X']
temp_yd = temp_Coor['Y']
temp_zd = temp_Coor['Z']
ax1.scatter3D(temp_xd, temp_yd, temp_zd, c=adata.uns['mclust_colors'][it],s=0.2, marker="o", label=label)
ax1.set_xlabel('')
ax1.set_ylabel('')
ax1.set_zlabel('')
ax1.set_xticklabels([])
ax1.set_yticklabels([])
ax1.set_zticklabels([])
plt.legend(bbox_to_anchor=(1.2,0.8), markerscale=10, frameon=False)
plt.title('STAGATE-3D')
ax1.elev = 45
ax1.azim = -20
plt.show()
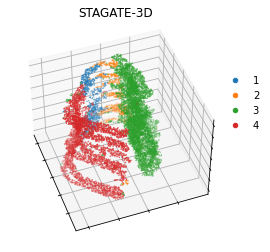
Running STAGATE with 2D spatial networks (for comparison)
The 2D spatial networks are saved at adata.uns (‘Spatial_Net_2D’). For camparison, we replace the 3D spatial networks with 2D spatial network and run STAGATE.
[20]:
adata_2D = adata.copy()
[21]:
adata_2D.uns['Spatial_Net'] = adata.uns['Spatial_Net_2D'].copy()
[22]:
adata_2D = STAGATE.train_STAGATE(adata_2D, alpha=0)
Size of Input: (10908, 3000)
2022-04-29 21:36:00.700666: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1639] Found device 0 with properties:
name: NVIDIA GeForce RTX 3090 major: 8 minor: 6 memoryClockRate(GHz): 1.695
pciBusID: 0000:65:00.0
2022-04-29 21:36:00.701352: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1639] Found device 1 with properties:
name: NVIDIA GeForce RTX 3090 major: 8 minor: 6 memoryClockRate(GHz): 1.695
pciBusID: 0000:b3:00.0
2022-04-29 21:36:00.701516: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcudart.so.10.0'; dlerror: libcudart.so.10.0: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701577: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcublas.so.10.0'; dlerror: libcublas.so.10.0: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701612: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcufft.so.10.0'; dlerror: libcufft.so.10.0: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701646: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcurand.so.10.0'; dlerror: libcurand.so.10.0: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701681: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcusolver.so.10.0'; dlerror: libcusolver.so.10.0: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701713: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcusparse.so.10.0'; dlerror: libcusparse.so.10.0: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701747: W tensorflow/stream_executor/platform/default/dso_loader.cc:55] Could not load dynamic library 'libcudnn.so.7'; dlerror: libcudnn.so.7: 无法打开共享对象文件: 没有那个文件或目录
2022-04-29 21:36:00.701752: W tensorflow/core/common_runtime/gpu/gpu_device.cc:1662] Cannot dlopen some GPU libraries. Please make sure the missing libraries mentioned above are installed properly if you would like to use GPU. Follow the guide at https://www.tensorflow.org/install/gpu for how to download and setup the required libraries for your platform.
Skipping registering GPU devices...
2022-04-29 21:36:00.701779: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1180] Device interconnect StreamExecutor with strength 1 edge matrix:
2022-04-29 21:36:00.701783: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1186] 0 1
2022-04-29 21:36:00.701787: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1199] 0: N N
2022-04-29 21:36:00.701789: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1199] 1: N N
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 500/500 [08:15<00:00, 1.01it/s]
[23]:
sc.pp.neighbors(adata_2D, use_rep='STAGATE')
sc.tl.umap(adata_2D)
[24]:
num_cluster = 4
adata_2D = STAGATE.mclust_R(adata_2D, num_cluster, used_obsm='STAGATE')
[25]:
adata_2D.uns['Section_id_colors'] = ['#02899A', '#0E994D', '#86C049', '#FBB21F', '#F48022', '#DA5326', '#BA3326']
[26]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.umap(adata_2D, color=['mclust', 'Section_id'])
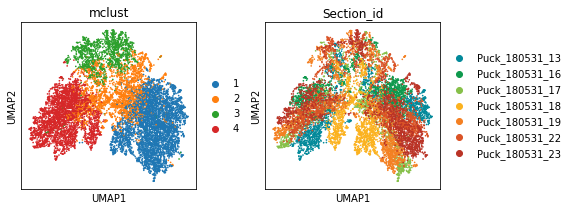
[27]:
fig = plt.figure(figsize=(4, 4))
ax1 = plt.axes(projection='3d')
for it, label in enumerate(np.unique(adata_2D.obs['mclust'])):
temp_Coor = adata_2D.obs.loc[adata_2D.obs['mclust']==label, :]
temp_xd = temp_Coor['X']
temp_yd = temp_Coor['Y']
temp_zd = temp_Coor['Z']
ax1.scatter3D(temp_xd, temp_yd, temp_zd, c=adata_2D.uns['mclust_colors'][it],s=0.2, marker="o", label=label)
ax1.set_xlabel('')
ax1.set_ylabel('')
ax1.set_zlabel('')
ax1.set_xticklabels([])
ax1.set_yticklabels([])
ax1.set_zticklabels([])
plt.legend(bbox_to_anchor=(1.2,0.8), markerscale=10, frameon=False)
plt.title('STAGATE-2D')
ax1.elev = 45
ax1.azim = -20
plt.show()
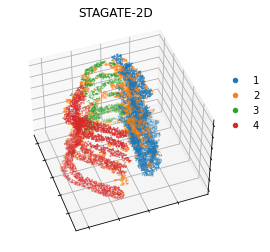
STAGATE-2D vs STAGATE-3D
[28]:
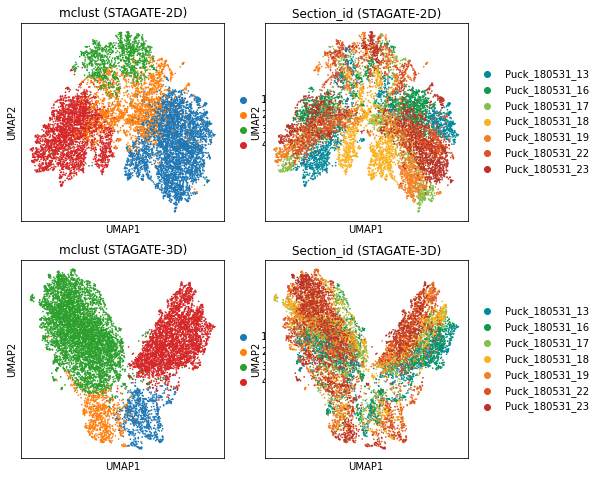
fig, axs = plt.subplots(2, 2, figsize=(8,8))
sc.pl.umap(adata_2D, color='mclust', title='mclust (STAGATE-2D)', show=False, ax=axs[0,0])
sc.pl.umap(adata_2D, color='Section_id', title='Section_id (STAGATE-2D)', show=False, ax=axs[0,1])
sc.pl.umap(adata, color='mclust', title='mclust (STAGATE-3D)', show=False, ax=axs[1,0])
sc.pl.umap(adata, color='Section_id', title='Section_id (STAGATE-3D)', show=False, ax=axs[1,1])
[28]:
<AxesSubplot:title={'center':'Section_id (STAGATE-3D)'}, xlabel='UMAP1', ylabel='UMAP2'>
[ ]: