Tutorial 1: 10x Visium (DLPFC dataset)
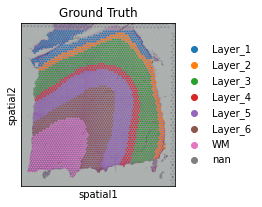
Here we present our re-analysis of 151676 sample of the dorsolateral prefrontal cortex (DLPFC) dataset. Maynard et al. has manually annotated DLPFC layers and white matter (WM) based on the morphological features and gene markers.
This tutorial demonstrates how to identify spatial domains on 10x Visium data using STAGATE. The processed data are available at https://github.com/LieberInstitute/spatialLIBD. We downloaded the manual annotation from the spatialLIBD package and provided at https://drive.google.com/drive/folders/10lhz5VY7YfvHrtV40MwaqLmWz56U9eBP?usp=sharing.
Preparation
[1]:
import warnings
warnings.filterwarnings("ignore")
[2]:
import pandas as pd
import numpy as np
import scanpy as sc
import matplotlib.pyplot as plt
import os
import sys
[3]:
from sklearn.metrics.cluster import adjusted_rand_score
[4]:
import STAGATE
[5]:
# the location of R (used for the mclust clustering)
os.environ['R_HOME'] = 'D:\Program Files\R\R-4.0.3'
os.environ['R_USER'] = 'D:\ProgramData\Anaconda3\Lib\site-packages\rpy2'
[6]:
section_id = '151676'
[7]:
input_dir = os.path.join('Data', section_id)
adata = sc.read_visium(path=input_dir, count_file=section_id+'_filtered_feature_bc_matrix.h5')
adata.var_names_make_unique()
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
[8]:
adata
[8]:
AnnData object with n_obs × n_vars = 3460 × 33538
obs: 'in_tissue', 'array_row', 'array_col'
var: 'gene_ids', 'feature_types', 'genome'
uns: 'spatial'
obsm: 'spatial'
[9]:
#Normalization
sc.pp.highly_variable_genes(adata, flavor="seurat_v3", n_top_genes=3000)
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
[10]:
# read the annotation
Ann_df = pd.read_csv(os.path.join('Data', section_id, section_id+'_truth.txt'), sep='\t', header=None, index_col=0)
Ann_df.columns = ['Ground Truth']
[11]:
adata.obs['Ground Truth'] = Ann_df.loc[adata.obs_names, 'Ground Truth']
[12]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.spatial(adata, img_key="hires", color=["Ground Truth"])
... storing 'Ground Truth' as categorical
... storing 'feature_types' as categorical
... storing 'genome' as categorical
Constructing the spatial network
[13]:
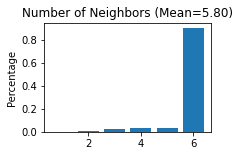
STAGATE.Cal_Spatial_Net(adata, rad_cutoff=150)
STAGATE.Stats_Spatial_Net(adata)
------Calculating spatial graph...
The graph contains 20052 edges, 3460 cells.
5.7954 neighbors per cell on average.
Running STAGATE
[14]:
adata = STAGATE.train_STAGATE(adata, alpha=0)
Size of Input: (3460, 3000)
WARNING:tensorflow:From D:\ProgramData\Anaconda3\lib\site-packages\tensorflow_core\python\ops\math_grad.py:1375: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.where in 2.0, which has the same broadcast rule as np.where
100%|████████████████████████████████████████████████████████████████████████████████| 500/500 [00:28<00:00, 17.60it/s]
[15]:
sc.pp.neighbors(adata, use_rep='STAGATE')
sc.tl.umap(adata)
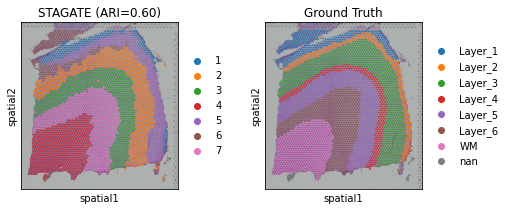
adata = STAGATE.mclust_R(adata, used_obsm='STAGATE', num_cluster=7)
[16]:
obs_df = adata.obs.dropna()
ARI = adjusted_rand_score(obs_df['mclust'], obs_df['Ground Truth'])
print('Adjusted rand index = %.2f' %ARI)
Adjusted rand index = 0.60
[17]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.umap(adata, color=["mclust", "Ground Truth"], title=['STAGATE (ARI=%.2f)'%ARI, "Ground Truth"])

[18]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.spatial(adata, color=["mclust", "Ground Truth"], title=['STAGATE (ARI=%.2f)'%ARI, "Ground Truth"])
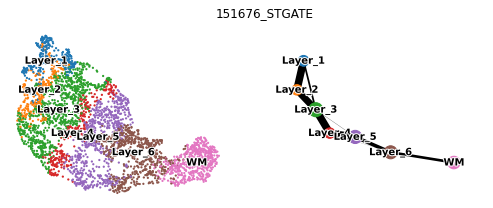
Spatial trajectory inference (PAGA)
[19]:
used_adata = adata[adata.obs['Ground Truth']!='nan',]
used_adata
[19]:
View of AnnData object with n_obs × n_vars = 3431 × 33538
obs: 'in_tissue', 'array_row', 'array_col', 'Ground Truth', 'mclust'
var: 'gene_ids', 'feature_types', 'genome', 'highly_variable', 'highly_variable_rank', 'means', 'variances', 'variances_norm'
uns: 'spatial', 'hvg', 'log1p', 'Ground Truth_colors', 'Spatial_Net', 'neighbors', 'umap', 'mclust_colors'
obsm: 'spatial', 'STAGATE', 'X_umap'
obsp: 'distances', 'connectivities'
[20]:
sc.tl.paga(used_adata, groups='Ground Truth')
Trying to set attribute `.uns` of view, copying.
[21]:
plt.rcParams["figure.figsize"] = (4,3)
sc.pl.paga_compare(used_adata, legend_fontsize=10, frameon=False, size=20,
title=section_id+'_STGATE', legend_fontoutline=2, show=False)
[21]:
[<matplotlib.axes._axes.Axes at 0x183007d1e48>,
<matplotlib.axes._axes.Axes at 0x18300411f98>]