Additional Tutorial 2: Work for multiple sections (STAGATE + Harmony)
Here we combine STAGATE and harmony algorithms to remove batch effects.
The mouse olfactory bulb data generated by different platform (Slide-seqV2 and Stereo-seq) are used in this tutorial. The detials of these two datasets can be found in Tutorial 3 and Tutorial 4.
Preparation
[1]:
import pandas as pd
import numpy as np
import scanpy as sc
import os
import sys
import matplotlib.pyplot as plt
import seaborn as sns
import gc
import warnings
warnings.filterwarnings("ignore")
[2]:
import STAGATE
Load Data
[3]:
adata_list = {}
Slide-seqV2
[4]:
input_dir = 'MOB_Data/Slide-seqV2'
counts_file = os.path.join(input_dir, 'Puck_200127_15.digital_expression.txt')
coor_file = os.path.join(input_dir, 'Puck_200127_15_bead_locations.csv')
[5]:
counts = pd.read_csv(counts_file, sep='\t', index_col=0)
coor_df = pd.read_csv(coor_file, index_col=0)
print(counts.shape, coor_df.shape)
(21220, 21724) (21724, 2)
[6]:
adata = sc.AnnData(counts.T)
adata.var_names_make_unique()
coor_df = coor_df.loc[adata.obs_names, ['xcoord', 'ycoord']]
adata.obsm["spatial"] = coor_df.to_numpy()
[7]:
sc.pp.calculate_qc_metrics(adata, inplace=True)
[8]:
adata
[8]:
AnnData object with n_obs × n_vars = 21724 × 21220
obs: 'n_genes_by_counts', 'log1p_n_genes_by_counts', 'total_counts', 'log1p_total_counts', 'pct_counts_in_top_50_genes', 'pct_counts_in_top_100_genes', 'pct_counts_in_top_200_genes', 'pct_counts_in_top_500_genes'
var: 'n_cells_by_counts', 'mean_counts', 'log1p_mean_counts', 'pct_dropout_by_counts', 'total_counts', 'log1p_total_counts'
obsm: 'spatial'
[9]:
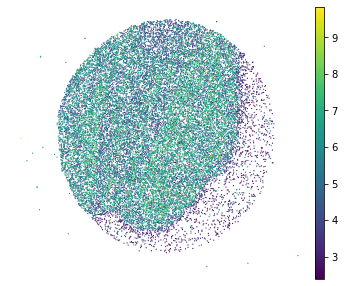
plt.rcParams["figure.figsize"] = (6,5)
sc.pl.embedding(adata, basis="spatial", color="log1p_total_counts",s=6, show=False)
plt.title('')
plt.axis('off')
[9]:
(-289.81710000000004, 6312.7151, 173.30850000000004, 5709.8615)
[10]:
# can be downloaded from https://drive.google.com/drive/folders/10lhz5VY7YfvHrtV40MwaqLmWz56U9eBP?usp=sharing
used_barcode = pd.read_csv(os.path.join(input_dir, 'used_barcodes.txt'), sep='\t', header=None)
used_barcode = used_barcode[0]
[11]:
adata = adata[used_barcode,]
adata
[11]:
View of AnnData object with n_obs × n_vars = 20139 × 21220
obs: 'n_genes_by_counts', 'log1p_n_genes_by_counts', 'total_counts', 'log1p_total_counts', 'pct_counts_in_top_50_genes', 'pct_counts_in_top_100_genes', 'pct_counts_in_top_200_genes', 'pct_counts_in_top_500_genes'
var: 'n_cells_by_counts', 'mean_counts', 'log1p_mean_counts', 'pct_dropout_by_counts', 'total_counts', 'log1p_total_counts'
obsm: 'spatial'
[12]:
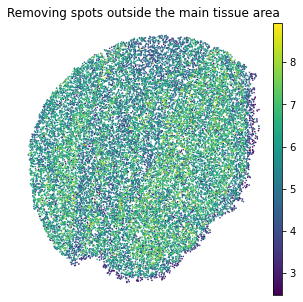
plt.rcParams["figure.figsize"] = (5,5)
sc.pl.embedding(adata, basis="spatial", color="log1p_total_counts",s=10, show=False, title='Removing spots outside the main tissue area')
plt.axis('off')
[12]:
(588.545, 5108.555, 847.6700000000001, 5670.73)
[13]:
sc.pp.filter_genes(adata, min_cells=50)
print('After flitering: ', adata.shape)
Trying to set attribute `.var` of view, copying.
After flitering: (20139, 11750)
[14]:
# make spot name unique
adata.obs_names = [x+'_SlideSeqV2' for x in adata.obs_names]
[15]:
adata_list['SlideSeqV2'] = adata.copy()
Stereo-seq
[16]:
input_dir = 'MOB_Data/Stereo-seq'
counts_file = os.path.join(input_dir, 'RNA_counts.tsv')
coor_file = os.path.join(input_dir, 'position.tsv')
[17]:
counts = pd.read_csv(counts_file, sep='\t', index_col=0)
coor_df = pd.read_csv(coor_file, sep='\t')
print(counts.shape, coor_df.shape)
(27106, 19527) (19527, 3)
[18]:
counts.columns = ['Spot_'+str(x) for x in counts.columns]
coor_df.index = coor_df['label'].map(lambda x: 'Spot_'+str(x))
coor_df = coor_df.loc[:, ['x','y']]
[19]:
coor_df.head()
[19]:
| x | y | |
|---|---|---|
| label | ||
| Spot_1 | 12555.007833 | 6307.537859 |
| Spot_2 | 12623.666667 | 6297.166667 |
| Spot_3 | 12589.567164 | 6302.552239 |
| Spot_4 | 12642.495050 | 6307.386139 |
| Spot_5 | 13003.333333 | 6307.990991 |
[20]:
adata = sc.AnnData(counts.T)
adata.var_names_make_unique()
[21]:
adata
[21]:
AnnData object with n_obs × n_vars = 19527 × 27106
[22]:
coor_df = coor_df.loc[adata.obs_names, ['y', 'x']]
adata.obsm["spatial"] = coor_df.to_numpy()
sc.pp.calculate_qc_metrics(adata, inplace=True)
[23]:
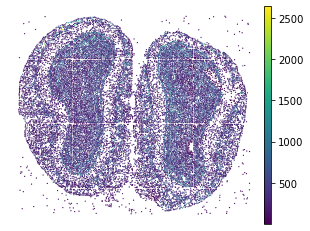
plt.rcParams["figure.figsize"] = (5,4)
sc.pl.embedding(adata, basis="spatial", color="n_genes_by_counts", show=False)
plt.title("")
plt.axis('off')
[23]:
(6002.432692307693, 12486.580128205129, 9908.545833333334, 15086.093055555555)
[24]:
used_barcode = pd.read_csv(os.path.join(input_dir, 'used_barcodes.txt'), sep='\t', header=None)
used_barcode = used_barcode[0]
adata = adata[used_barcode,]
[25]:
adata
[25]:
View of AnnData object with n_obs × n_vars = 19109 × 27106
obs: 'n_genes_by_counts', 'log1p_n_genes_by_counts', 'total_counts', 'log1p_total_counts', 'pct_counts_in_top_50_genes', 'pct_counts_in_top_100_genes', 'pct_counts_in_top_200_genes', 'pct_counts_in_top_500_genes'
var: 'n_cells_by_counts', 'mean_counts', 'log1p_mean_counts', 'pct_dropout_by_counts', 'total_counts', 'log1p_total_counts'
obsm: 'spatial'
[26]:
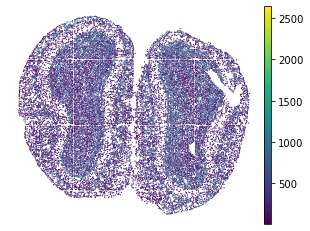
plt.rcParams["figure.figsize"] = (5,4)
sc.pl.embedding(adata, basis="spatial", color="n_genes_by_counts", show=False)
plt.title("")
plt.axis('off')
[26]:
(6005.190789473685, 12428.6600877193, 9986.774763741741, 15062.302776957436)
[27]:
sc.pp.filter_genes(adata, min_cells=50)
print('After flitering: ', adata.shape)
Trying to set attribute `.var` of view, copying.
After flitering: (19109, 14376)
[28]:
# make spot name unique
adata.obs_names = [x+'_StereoSeq' for x in adata.obs_names]
[29]:
adata_list['StereoSeq'] = adata.copy()
Constructing the spatial network for each secion
Slide-seqV2
[30]:
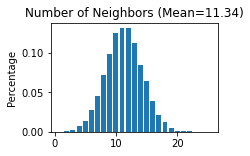
STAGATE.Cal_Spatial_Net(adata_list['SlideSeqV2'], rad_cutoff=50)
STAGATE.Stats_Spatial_Net(adata_list['SlideSeqV2'])
------Calculating spatial graph...
The graph contains 228300 edges, 20139 cells.
11.3362 neighbors per cell on average.
[31]:
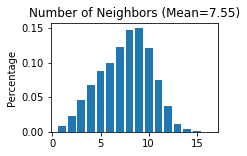
STAGATE.Cal_Spatial_Net(adata_list['StereoSeq'], rad_cutoff=50)
STAGATE.Stats_Spatial_Net(adata_list['StereoSeq'])
------Calculating spatial graph...
The graph contains 144318 edges, 19109 cells.
7.5524 neighbors per cell on average.
Note that the spatial network are saved in adata.uns[‘Spatial_Net’], which can be conbat directly for multiple sections.
[32]:
adata_list['SlideSeqV2'].uns['Spatial_Net']
[32]:
| Cell1 | Cell2 | Distance | |
|---|---|---|---|
| 0 | AAAAAAACAAAAGG_SlideSeqV2 | ATAAGTTGCCCCGT_SlideSeqV2 | 41.494698 |
| 1 | AAAAAAACAAAAGG_SlideSeqV2 | CTCCGGGCTCTTCA_SlideSeqV2 | 44.777226 |
| 2 | AAAAAAACAAAAGG_SlideSeqV2 | CCAGCAAAGCTACA_SlideSeqV2 | 29.429237 |
| 3 | AAAAAAACAAAAGG_SlideSeqV2 | GCCTAAAGCTTTTG_SlideSeqV2 | 25.927784 |
| 5 | AAAAAAACAAAAGG_SlideSeqV2 | CCTCCTTAACGTTA_SlideSeqV2 | 33.634060 |
| ... | ... | ... | ... |
| 9 | TTTTTTTTTTTTAT_SlideSeqV2 | CCTATAACAGCCTG_SlideSeqV2 | 30.802922 |
| 10 | TTTTTTTTTTTTAT_SlideSeqV2 | CTTGGGCATATAAG_SlideSeqV2 | 37.316216 |
| 11 | TTTTTTTTTTTTAT_SlideSeqV2 | AGTAGTTGCGGCCG_SlideSeqV2 | 13.688316 |
| 12 | TTTTTTTTTTTTAT_SlideSeqV2 | TGTATTCACTTTGC_SlideSeqV2 | 18.753666 |
| 13 | TTTTTTTTTTTTAT_SlideSeqV2 | TAAAACGCGCGAGA_SlideSeqV2 | 25.347584 |
228300 rows × 3 columns
Conbat the scanpy objects and spatial networks
[33]:
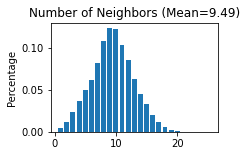
adata = sc.concat([adata_list['SlideSeqV2'], adata_list['StereoSeq']], keys=None)
[34]:
adata.uns['Spatial_Net'] = pd.concat([adata_list['SlideSeqV2'].uns['Spatial_Net'], adata_list['StereoSeq'].uns['Spatial_Net']])
[35]:
STAGATE.Stats_Spatial_Net(adata)
Normalization
[36]:
#Normalization
sc.pp.highly_variable_genes(adata, flavor="seurat_v3", n_top_genes=3000)
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
Running STAGATE
[37]:
adata = STAGATE.train_STAGATE(adata, alpha=0)
Size of Input: (39248, 3000)
WARNING:tensorflow:From /home/dkn/anaconda3/envs/STAGATE_tf/lib/python3.6/site-packages/tensorflow_core/python/ops/math_grad.py:1375: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.where in 2.0, which has the same broadcast rule as np.where
100%|██████████| 500/500 [18:06<00:00, 2.17s/it]
[38]:
sc.pp.neighbors(adata, use_rep='STAGATE')
sc.tl.umap(adata)
[39]:
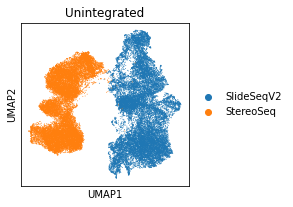
adata.obs['Tech'] = [x.split('_')[-1] for x in adata.obs_names]
[40]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.umap(adata, color='Tech', title='Unintegrated')
... storing 'Tech' as categorical
Run Harmony on the STAGATE represention
[41]:
import harmonypy as hm
[42]:
data_mat = adata.obsm['STAGATE'].copy()
meta_data = adata.obs.copy()
[43]:
# Run Harmony
ho = hm.run_harmony(data_mat, meta_data, ['Tech'])
2022-10-30 18:54:54,390 - harmonypy - INFO - Iteration 1 of 10
2022-10-30 18:55:05,947 - harmonypy - INFO - Iteration 2 of 10
2022-10-30 18:55:20,272 - harmonypy - INFO - Iteration 3 of 10
2022-10-30 18:55:24,721 - harmonypy - INFO - Converged after 3 iterations
[44]:
# Write the adjusted PCs to a new file.
res = pd.DataFrame(ho.Z_corr)
res.columns = adata.obs_names
[45]:
adata_Harmony = sc.AnnData(res.T)
[46]:
adata_Harmony.obsm['spatial'] = pd.DataFrame(adata.obsm['spatial'], index=adata.obs_names).loc[adata_Harmony.obs_names,].values
adata_Harmony.obs['Tech'] = adata.obs.loc[adata_Harmony.obs_names, 'Tech']
[47]:
sc.pp.neighbors(adata_Harmony)
sc.tl.umap(adata_Harmony)
[48]:
sc.tl.louvain(adata_Harmony, resolution=0.8)
[49]:
plt.rcParams["figure.figsize"] = (3, 3)
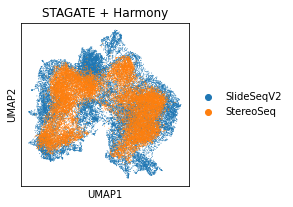
sc.pl.umap(adata_Harmony, color='Tech', title='STAGATE + Harmony')
[50]:
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.umap(adata_Harmony, color='louvain')

[ ]:
[60]:
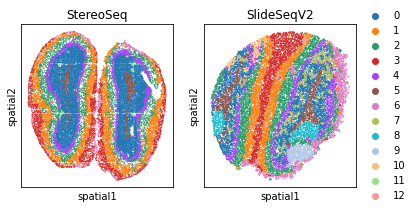
fig, axs = plt.subplots(1, 2, figsize=(6, 3))
it=0
for temp_tech in ['StereoSeq', 'SlideSeqV2']:
temp_adata = adata_Harmony[adata_Harmony.obs['Tech']==temp_tech, ]
if it == 1:
sc.pl.embedding(temp_adata, basis="spatial", color="louvain",s=6, ax=axs[it],
show=False, title=temp_tech)
else:
sc.pl.embedding(temp_adata, basis="spatial", color="louvain",s=6, ax=axs[it], legend_loc=None,
show=False, title=temp_tech)
it+=1
[ ]: